Selected Publications
CRISPR-induced DNA reorganization for multiplexed nucleic acid detection
Karlikow M, Amalfitano E, Yang X, Doucet J, Chapman A, Mousavi PS, Homme P, Sutyrina P, Chan W, Lemak S, Yakunin AF, Dolezal AG, Kelley S, Foster LJ, Harpur BA, Pardee K. CRISPR-induced DNA reorganization for multiplexed nucleic acid detection. Nature Communications. 2023 Mar 17.
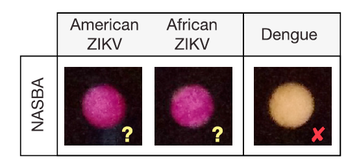
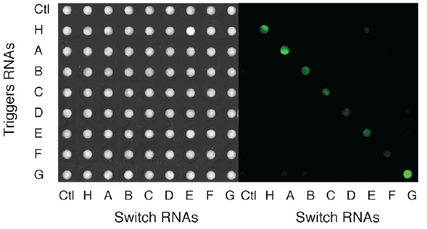
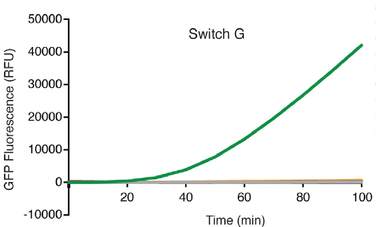
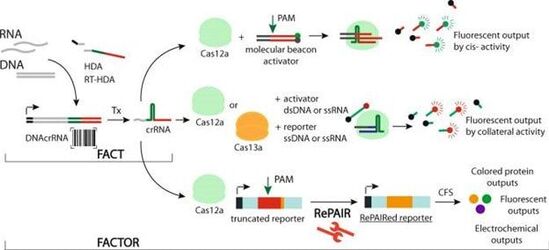
Nucleic acid sensing powered by the sequence recognition of CRIPSR technologies has enabled major advancement toward rapid, accurate and deployable diagnostics. While exciting, there are still many challenges facing their practical implementation, such as the widespread need for a PAM sequence in the targeted nucleic acid, labile RNA inputs, and limited multiplexing. Here we report FACT (Functionalized Amplification CRISPR Tracing), a CRISPR-based nucleic acid barcoding technology compatible with Cas12a and Cas13a, enabling diagnostic outputs based on cis- and trans-cleavage from any sequence. Furthermore, we link the activation of CRISPR-Cas12a to the expression of proteins through a Reprogrammable PAIRing system (RePAIR). We then combine FACT and RePAIR to create FACTOR (FACT on RePAIR), a CRISPR-based diagnostic, that we use to detect infectious disease in an agricultural use case: honey bee viral infection. With high specificity and accuracy, we demonstrate the potential of FACTOR to be applied to the sensing of any nucleic acid of interest.
Karlikow M, Amalfitano E, Yang X, Doucet J, Chapman A, Mousavi PS, Homme P, Sutyrina P, Chan W, Lemak S, Yakunin AF, Dolezal AG, Kelley S, Foster LJ, Harpur BA, Pardee K. CRISPR-induced DNA reorganization for multiplexed nucleic acid detection. Nature Communications. 2023 Mar 17.
Nucleic acid sensing powered by the sequence recognition of CRIPSR technologies has enabled major advancement toward rapid, accurate and deployable diagnostics. While exciting, there are still many challenges facing their practical implementation, such as the widespread need for a PAM sequence in the targeted nucleic acid, labile RNA inputs, and limited multiplexing. Here we report FACT (Functionalized Amplification CRISPR Tracing), a CRISPR-based nucleic acid barcoding technology compatible with Cas12a and Cas13a, enabling diagnostic outputs based on cis- and trans-cleavage from any sequence. Furthermore, we link the activation of CRISPR-Cas12a to the expression of proteins through a Reprogrammable PAIRing system (RePAIR). We then combine FACT and RePAIR to create FACTOR (FACT on RePAIR), a CRISPR-based diagnostic, that we use to detect infectious disease in an agricultural use case: honey bee viral infection. With high specificity and accuracy, we demonstrate the potential of FACTOR to be applied to the sensing of any nucleic acid of interest.
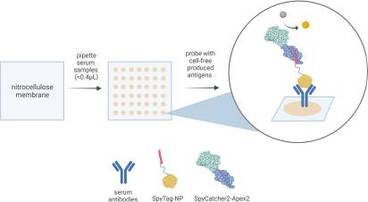
Cell-Free Dot Blot: an ultra-low-cost and practical immunoassay platform for detection of anti-SARS-CoV-2 antibodies in human and animal sera
Norouzi M, Truong T, Jaenes K, Warner BM, Vendramelli R, Tierney K, Kobasa D, Tailor N, Plant P, Dos Santos C, Babiuk S, Ambagala A, Pardee K. Cell-Free Dot Blot: an Ultra-Low-Cost and Practical Immunoassay Platform for Detection of Anti-SARS-CoV-2 Antibodies in Human and Animal Sera. Microbiology Spectrum. 2023 Mar/Apr
Since its emergence in late 2019, the coronavirus disease 2019 (COVID-19) pandemic has caused severe disruption to key aspects of human life globally and highlighted the need for timely, adaptive, and accessible pandemic response strategies. Here, we introduce the cell-free dot blot (CFDB) method, a practical and ultra-low-cost immune diagnostic platform capable of rapid response and mass immunity screening for the current and future pandemics. Similar in mechanism to the widely used enzyme-linked immunosorbent assays (ELISAs), our method is novel and advantageous in that (i) it uses linear DNA to produce the target viral antigen fused to a SpyTag peptide in a cell-free expression system without the need for traditional cloning and antigen purification, (ii) it uses SpyCatcher2-Apex2, an Escherichia coli-produced peroxidase conjugate as a universal secondary detection reagent, obviating the need for commercial or sophisticated enzyme conjugates, and (iii) sera are spotted directly on a nitrocellulose membrane, enabling a simple “dipping” mechanism for downstream incubation and washing steps, as opposed to individual processing of wells in a multiwell plate. To demonstrate the utility of our method, we performed CFDB to detect anti-severe acute respiratory syndrome coronavirus 2 nucleocapsid protein antibodies in precharacterized human sera (23 negative and 36 positive for COVID-19) and hamster sera (16 negative and 36 positive for COVID-19), including independent testing at a collaborating laboratory, and we show assay performance comparable to that of conventional ELISAs. At a similar capacity to 96-well plate ELISA kits, one CFDB assay costs only ~$3 USD. We believe that CFDB can become a valuable pandemic response tool for adaptive and accessible sero-surveillance in human and animal populations.
Norouzi M, Truong T, Jaenes K, Warner BM, Vendramelli R, Tierney K, Kobasa D, Tailor N, Plant P, Dos Santos C, Babiuk S, Ambagala A, Pardee K. Cell-Free Dot Blot: an Ultra-Low-Cost and Practical Immunoassay Platform for Detection of Anti-SARS-CoV-2 Antibodies in Human and Animal Sera. Microbiology Spectrum. 2023 Mar/Apr
Since its emergence in late 2019, the coronavirus disease 2019 (COVID-19) pandemic has caused severe disruption to key aspects of human life globally and highlighted the need for timely, adaptive, and accessible pandemic response strategies. Here, we introduce the cell-free dot blot (CFDB) method, a practical and ultra-low-cost immune diagnostic platform capable of rapid response and mass immunity screening for the current and future pandemics. Similar in mechanism to the widely used enzyme-linked immunosorbent assays (ELISAs), our method is novel and advantageous in that (i) it uses linear DNA to produce the target viral antigen fused to a SpyTag peptide in a cell-free expression system without the need for traditional cloning and antigen purification, (ii) it uses SpyCatcher2-Apex2, an Escherichia coli-produced peroxidase conjugate as a universal secondary detection reagent, obviating the need for commercial or sophisticated enzyme conjugates, and (iii) sera are spotted directly on a nitrocellulose membrane, enabling a simple “dipping” mechanism for downstream incubation and washing steps, as opposed to individual processing of wells in a multiwell plate. To demonstrate the utility of our method, we performed CFDB to detect anti-severe acute respiratory syndrome coronavirus 2 nucleocapsid protein antibodies in precharacterized human sera (23 negative and 36 positive for COVID-19) and hamster sera (16 negative and 36 positive for COVID-19), including independent testing at a collaborating laboratory, and we show assay performance comparable to that of conventional ELISAs. At a similar capacity to 96-well plate ELISA kits, one CFDB assay costs only ~$3 USD. We believe that CFDB can become a valuable pandemic response tool for adaptive and accessible sero-surveillance in human and animal populations.
Field Validation of the Performance of Paper-Based Tests for the Detection of the Zika And Chikungunya Viruses in Serum Samples
Karlikow M, Ribeiro J, Guo Y, Cicek S, Krokovski L, Homme P, Xiong Y, Xu T, Calderon M, Camacho S, Ma D, Nayane B, Sutyrina P, Ferrante T, Benitez D, Tamayo V, Jaenes K, Rackus D, Collins J, Castellanos J, Cevallos V, Green AA, Ayres C, Pena L, Pardee K.
Nature Biomedical Engineering (March 2022)
Health security has implications that are far reaching and affect both well-being and economic stability. The recent Zika virus outbreak in Latin America highlighted these issues, especially in low-resource settings, where resilience to infectious disease outbreaks can be limited by access to health care tools. Here we report a field-based, patient trial in Latin America for a paper-based Zika diagnostic using cell-free expression systems, a purpose-built companion reader called PLUM and its onboard computer vision-enabled analysis software. Based on two sequence-specific steps, isothermal amplification and toehold switch-based sensors, we demonstrate sensitivity for target RNA sequences well within the clinically relevant range (2 attomolar). Using cultured virus, we then show analytical specificity and sensitivity equivalent to RT-qPCR for the Zika virus, and a diagnostic accuracy of 98.5% with 268 patient samples. We also achieved similar diagnostic performance for the chikungunya virus, demonstrating the approach’s programmability and extensibility. This work, on-site in Latin America, reveals the utility of cell-free synthetic biology tools and companion hardware for providing de-centralized, high-capacity, and low-cost diagnostics for use in low-resource settings.
Karlikow M, Ribeiro J, Guo Y, Cicek S, Krokovski L, Homme P, Xiong Y, Xu T, Calderon M, Camacho S, Ma D, Nayane B, Sutyrina P, Ferrante T, Benitez D, Tamayo V, Jaenes K, Rackus D, Collins J, Castellanos J, Cevallos V, Green AA, Ayres C, Pena L, Pardee K.
Nature Biomedical Engineering (March 2022)
Health security has implications that are far reaching and affect both well-being and economic stability. The recent Zika virus outbreak in Latin America highlighted these issues, especially in low-resource settings, where resilience to infectious disease outbreaks can be limited by access to health care tools. Here we report a field-based, patient trial in Latin America for a paper-based Zika diagnostic using cell-free expression systems, a purpose-built companion reader called PLUM and its onboard computer vision-enabled analysis software. Based on two sequence-specific steps, isothermal amplification and toehold switch-based sensors, we demonstrate sensitivity for target RNA sequences well within the clinically relevant range (2 attomolar). Using cultured virus, we then show analytical specificity and sensitivity equivalent to RT-qPCR for the Zika virus, and a diagnostic accuracy of 98.5% with 268 patient samples. We also achieved similar diagnostic performance for the chikungunya virus, demonstrating the approach’s programmability and extensibility. This work, on-site in Latin America, reveals the utility of cell-free synthetic biology tools and companion hardware for providing de-centralized, high-capacity, and low-cost diagnostics for use in low-resource settings.
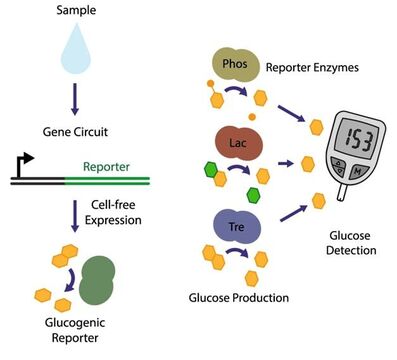
A Glucose Meter Interface for Point-Of-Care Gene Circuit-Based Diagnostics
Amalfitano E, Karlikow M, Norouzi M, Jaenes K, Cicek S, Masum F, Sadat Mousavi P, Guo Y, Tang L, Sydor A, Ma D, Pearson JD, Trcka D, Pinette M, Ambagala A, Babiuk S, Pickering B, Wrana J, Bremner R, Mazzulli T, Sinton D, Brumell JH, Green AA, Pardee K.
Nature Communications. 2021 Feb 1.
This project aims to solve the challenge of “How to practically deploy gene circuit-based sensors into the existing diagnostic ecosystem?”. The blood glucose monitor is arguably the most widely used diagnostic device and has “revolutionized” the lives of millions of diabetics by enabling the portable quantification of blood sugar, and therefore personal management. In this publication, we reported the development of a molecular “translator” that converts the output from our gene circuit-based diagnostics into one that is compatible with an off-the-shelf glucose monitor. This is done through the expression of a reporter enzyme that generates glucose from otherwise inert glucose-containing substrates (e.g. lactose, cellulose, starch). Once activated, the diagnostic sensor generates an enzyme to degrade these polymer substrates into monomeric glucose.
Proof-of-concept demonstrations included detection of RNA sequences from Typhoid, Paratyphoid A and B, and related drug resistance genes. Follow-on work found that the glucose meter interface could detect as few as 1000 colony forming units of S. typhi per mL, which is well-within the detection range of the current standard lab-based Typhoid diagnostic method, blood culture (1000 – 43,500 DNA copies/mL).
With the onset of the COVID-19 pandemic, we adapted the glucose meter interface for the detection of SARS-CoV-2 using the toehold switch-based sensors for gene N and gene E from above. Using samples provided by the National Microbiology Laboratory and Mt. Sinai hospital, the resulting glucose meter-based system was able to detect the viral genome at clinically relevant concentrations down to 16 aM and, in a small test with patient samples, could distinguish between COVID-19+ patients and healthy volunteers.
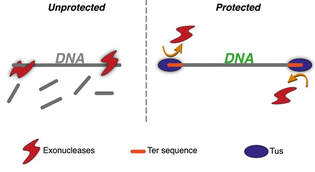
High-Efficiency Protection of Linear DNA in Cell-Free Extracts from Escherichia coli and Vibrio natriegens
Norouzi M, Panfilov S, Pardee K.
ACS Synthetic Biology. 2021 Jul 16;10(7):1615-1624.
Cell-free protein expression provide significant advantages (e.g. time, cost) over cell-based expression; however, key challenges remain to be solved. One such benefit is that, in the absence of a cell-wall, protein expression can be done rapidly by simply pipetting template DNA into reactions. This is generally performed using circular plasmid DNA templates because lysate-based cell-free systems contain exonucleases that degrade linear DNA (minutes). However, if linear DNA templates could be used directly in cell-free systems, this would save the several days needed for cell-based cloning the gene of interest into a plasmid.
Recognizing the importance of this challenge, we developed a method, based on the Tus-Ter E. coli DNA replication termination system, that provides protection to linear DNA for highly efficient cell-free protein expression. The system works by adding a short Ter sequence (23 bp) to DNA constructs during commercial gene synthesis or as a primer overhang using PCR. The high-affinity binding of Tus to the Ter sequence (KD = 3.4 x 10-13 M) strongly inhibits the progress of helicase-containing nucleases toward any DNA sequence preceding the Ter site. In proof-of-concept work, we showed that the Tus-Ter system enables cell-free expression from linear DNA templates at yields equal to or higher than plasmid DNA in lysates from both E. coli and Vibrio natriegens.
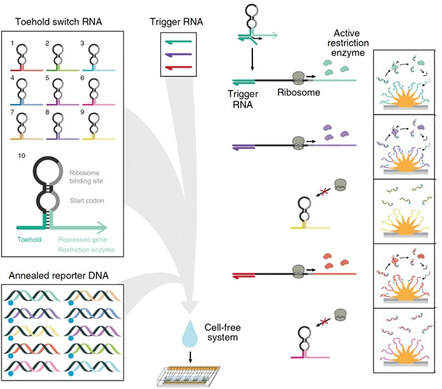
A Multiplexed, Electrochemical Interface for Gene Circuit-Based Sensors
Sadat MP, Smith SJ, Chen JB, Karlikow M, Tinafar A, Robinson C, Liu W, Ma D, Green AA, Kelley SO, Pardee K.
Nature Chemistry. 2020 Jan;12(1):48-55.
The components used in the field of synthetic biology, including gene circuits, are often described using analogous terms from electronics (e.g. circuit, toggle switch, oscillator, logic gate, memory). However, thus far, these features have had to be laboriously encoded into the gene circuits themselves, which can take months to years. If synthetic gene networks could be interfaced directly with electronics, the burden of such logic operations could be off-loaded to electronic devices. Electronic integration would allow for flexible and on-the-fly changes to logic terms, and for seamless sharing of biosensor data.
With this vision in mind, and in collaboration with the Kelley lab, we have developed the first direct gene circuit/electrode interface that allows for output signals from gene circuit-based sensors to be transmitted directly to electrodes. In this work, we developed a scalable system of reporter enzymes that cleave specific DNA sequences in solution, which, in turn, create an electrochemical signal when these newly liberated strands are captured at the surface of multiplexed, nanostructured microelectrodes. In this publication we describe the development of this molecular-electrochemical interface and demonstrate its utility using a ligand-inducible gene circuit and toehold switch-based sensors, including the detection of multiple antibiotic resistance genes in parallel.
Sadat MP, Smith SJ, Chen JB, Karlikow M, Tinafar A, Robinson C, Liu W, Ma D, Green AA, Kelley SO, Pardee K.
Nature Chemistry. 2020 Jan;12(1):48-55.
The components used in the field of synthetic biology, including gene circuits, are often described using analogous terms from electronics (e.g. circuit, toggle switch, oscillator, logic gate, memory). However, thus far, these features have had to be laboriously encoded into the gene circuits themselves, which can take months to years. If synthetic gene networks could be interfaced directly with electronics, the burden of such logic operations could be off-loaded to electronic devices. Electronic integration would allow for flexible and on-the-fly changes to logic terms, and for seamless sharing of biosensor data.
With this vision in mind, and in collaboration with the Kelley lab, we have developed the first direct gene circuit/electrode interface that allows for output signals from gene circuit-based sensors to be transmitted directly to electrodes. In this work, we developed a scalable system of reporter enzymes that cleave specific DNA sequences in solution, which, in turn, create an electrochemical signal when these newly liberated strands are captured at the surface of multiplexed, nanostructured microelectrodes. In this publication we describe the development of this molecular-electrochemical interface and demonstrate its utility using a ligand-inducible gene circuit and toehold switch-based sensors, including the detection of multiple antibiotic resistance genes in parallel.
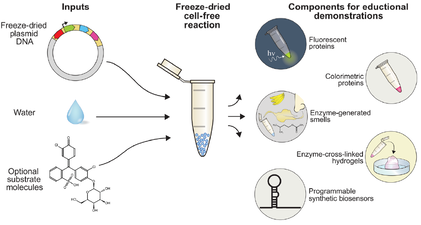
BioBits™ Explorer: A modular synthetic biology education kit
Huang A, Nguyen PQ, Stark JC, et al. BioBitsTM Explorer: A modular synthetic biology education kit. Science Advances. 2018;4(8):eaat5105. doi:10.1126/sciadv.aat5105.
Hands-on demonstrations greatly enhance the teaching of science, technology, engineering, and mathematics (STEM) concepts and foster engagement and exploration in the sciences. While numerous chemistry and physics classroom demonstrations exist, few biology demonstrations are practical and accessible due to the challengesand concerns of growing living cells in classrooms. We introduce BioBits™ Explorer, a synthetic biology educational kit based on shelf-stable, freeze-dried, cell-free (FD-CF) reactions, which are activated by simply adding water. The FD-CF reactions engage the senses of sight, smell, and touch with outputs that produce fluorescence, fragrances, and hydrogels, respectively. We introduce components that can teach tunable protein expression, enzymatic reactions, biomaterial formation, and biosensors using RNA switches, some of which represent original FD-CF outputs that expand the toolbox of cell-free synthetic biology. The BioBits™ Explorer kit enables hands-on demonstrations of cutting-edge science that are inexpensive and easy to use, circumventing many current barriers for implementing exploratory biology experiments in classrooms.
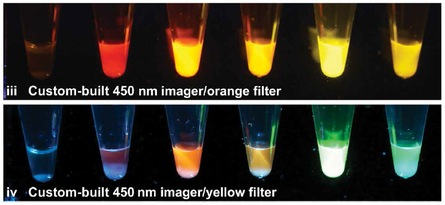
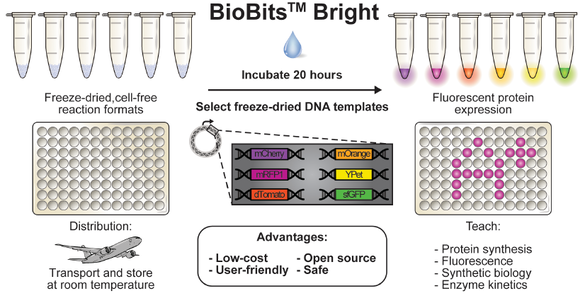
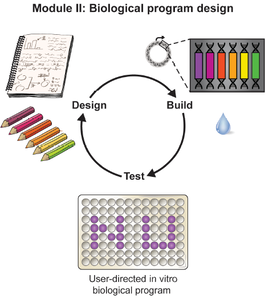
BioBits™ Bright: A fluorescent synthetic biology education kit
Stark JC, Huang A, Nguyen PQ, et al. BioBitsTM Bright: A fluorescent synthetic biology education kit. Science Advances. 2018;4(8):eaat5107. doi:10.1126/sciadv.aat5107.
Synthetic biology offers opportunities for experiential educational activities at the intersection of the life sciences, engineering, and design. However, implementation of hands-on biology activities in classrooms is challenging because of the need for specialized equipment and expertise to grow living cells. We present BioBits™ Bright, a shelfstable, just-add-water synthetic biology education kit with easy visual outputs enabled by expression of fluorescent proteins in freeze-dried, cell-free reactions. We introduce activities and supporting curricula for teaching the central dogma, tunable protein expression, and design-build-test cycles and report data generated by K-12 teachers and students. We also develop inexpensive incubators and imagers, resulting in a comprehensive kit costing <US$100 per 30-person classroom. The user-friendly resources of this kit promise to enhance biology education both inside and outside the classroom.
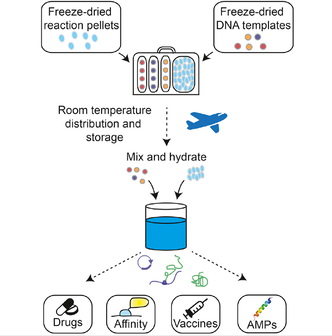
Portable, On-Demand Biomolecular Manufacturing.
Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, Ferrante T, McSorley FR, Furuta Y, Vernet A, Lewandowski M, Boddy CN, Joshi NS, Collins JJ Cell. 2016 Sep 22;167(1):248-259.e12. doi: 0.1016/j.cell.2016.09.013.
Synthetic biology uses living cells as molecular foundries for the biosynthesis of drugs, therapeutic proteins, and other commodities. However, the need for specialized equipment and refrigeration for production and distribution poses a challenge for the delivery of these technologies to the field and to low-resource areas. Here, we present a portable platform that provides the means for on-site, on-demand manufacturing of therapeutics and biomolecules. This flexible system is based on reaction pellets composed of freeze-dried, cell-free transcription and translation machinery, which can be easily hydrated and utilized for biosynthesis through the addition of DNA encoding the desired output. We demonstrate this approach with the manufacture and functional validation of antimicrobial peptides and vaccines and present combinatorial methods for the production of antibody conjugates and small molecules. This synthetic biology platform resolves important practical limitations in the production and distribution of therapeutics and molecular tools, both to the developed and developing world.
Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, Ferrante T, McSorley FR, Furuta Y, Vernet A, Lewandowski M, Boddy CN, Joshi NS, Collins JJ Cell. 2016 Sep 22;167(1):248-259.e12. doi: 0.1016/j.cell.2016.09.013.
Synthetic biology uses living cells as molecular foundries for the biosynthesis of drugs, therapeutic proteins, and other commodities. However, the need for specialized equipment and refrigeration for production and distribution poses a challenge for the delivery of these technologies to the field and to low-resource areas. Here, we present a portable platform that provides the means for on-site, on-demand manufacturing of therapeutics and biomolecules. This flexible system is based on reaction pellets composed of freeze-dried, cell-free transcription and translation machinery, which can be easily hydrated and utilized for biosynthesis through the addition of DNA encoding the desired output. We demonstrate this approach with the manufacture and functional validation of antimicrobial peptides and vaccines and present combinatorial methods for the production of antibody conjugates and small molecules. This synthetic biology platform resolves important practical limitations in the production and distribution of therapeutics and molecular tools, both to the developed and developing world.
Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components.
Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. Cell. 2016 May 19;165(5):1255-66. doi: 10.1016/j.cell.2016.04.059. Epub 2016 May 6.
Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. Cell. 2016 May 19;165(5):1255-66. doi: 10.1016/j.cell.2016.04.059. Epub 2016 May 6.
The recent Zika virus outbreak highlights the need for low-cost diagnostics that can be rapidly developed for distribution and use in pandemic regions. Here, we report a pipeline for the rapid design, assembly, and validation of cell-free, paper-based sensors for the detection of the Zika virus RNA genome. By linking isothermal RNA amplification to toehold switch RNA sensors, we detect clinically relevant concentrations of Zika virus sequences and demonstrate specificity against closely related Dengue virus sequences. When coupled with a novel CRISPR/Cas9-based module, our sensors can discriminate between viral strains with single-base resolution. We successfully demonstrate a simple, field-ready sample-processing workflow and detect Zika virus from the plasma of a viremic macaque. Our freeze-dried biomolecular platform resolves important practical limitations to the deployment of molecular diagnostics in the field and demonstrates how synthetic biology can be used to develop diagnostic tools for confronting global health crises.
Paper-based Synthetic Gene Networks.
Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. Cell. 2014 Nov 6;159(4):940-54. doi: 10.1016/j.cell.2014.10.004. Epub 2014 Oct 23.
Synthetic gene networks have wide-ranging uses in reprogramming and rewiring organisms. To date, there has not been a way to harness the vast potential of these networks beyond the constraints of a laboratory or in vivo environment. Here, we present an in vitro paper-based platform that provides an alternate, versatile venue for synthetic biologists to operate and a much-needed medium for the safe deployment of engineered gene circuits beyond the lab. Commercially available cell-free systems are freeze dried onto paper, enabling the inexpensive, sterile, and abiotic distribution of synthetic-biology-based technologies for the clinic, global health, industry, research, and education. For field use, we create circuits with colorimetric outputs for detection by eye and fabricate a low-cost, electronic optical interface. We demonstrate this technology with small-molecule and RNA actuation of genetic switches, rapid prototyping of complex gene circuits, and programmable in vitro diagnostics, including glucose sensors and strain-specific Ebola virus sensors.
All Publications
CRISPR-induced DNA reorganization for multiplexed nucleic acid detection
Karlikow M, Amalfitano E, Yang X, Doucet J, Chapman A, Mousavi PS, Homme P, Sutyrina P, Chan W, Lemak S, Yakunin AF, Dolezal AG, Kelley S, Foster LJ, Harpur B, Pardee K. Nature Communications. 2023 Mar 17;14(1):1505. PMID: 36932065.
Clinical and Laboratory Diagnosis of Monkeypox (Mpox): Current Status and Future Directions
Ribeiro da Silva J, Kohl A, Pena L, Pardee K. iScience 2023. 2023 Jun 16;26(6):106759. PMID: 37206155.
Cell-Free Dot Blot: an Ultra-Low-Cost and Practical Immunoassay Platform for Detection of Anti-SARS-CoV-2 Antibodies in Human and Animal Sera
Norouzi M, Truong T, Jaenes K, Warner BM, Vendramelli R, Tierney K, Kobasa D, Tailor N, Plant P, Dos Santos C, Babiuk S, Ambagala A, Pardee K. Microbiol Spectr 2023 Jan 31:e0245722. PMID: 36719206.
Recent insights into SARS-CoV-2 omicron variant
Ribeiro da Silva J, Kohl A, Pena L, Pardee K. Reviews in Medical Virology. e2373. (2023) PMID: 35662313
Field Validation of the Performance of Paper-Based Tests for the Detection of the Zika And Chikungunya Viruses in Serum Samples
Karlikow M, Ribeiro J, Guo Y, Cicek S, Krokovski L, Homme P, Xiong Y, Xu T, Calderon M, Camacho S, Ma D, Nayane B, Sutyrina P, Ferrante T, Benitez D, Tamayo V, Jaenes K, Rackus D, Collins J, Castellanos J, Cevallos V, Green AA, Ayres C, Pena L, Pardee K. Nature Biomedical Engineering, Mar 2022.
Toward Mail-in-Sensors for SARS-CoV-2 Detection: Interfacing Gel Switch Resonators with Cell-Free Toehold Switches
Carr AR, Dopp JL, Wu K, Sadat Mousavi P, Jo YR, McNeley CE, Lynch ZT, Pardee K, Green AA, Reuel NF. T. ACS Sensors. 7, 3, 806–815 (2022) PMID. 35254055.
From Design to Implementation: The Development and Patient Validation of Paper-Based Toehold Switch-Based Diagnostics
Jaenes K, Ribeiro J, Vigar J, Wu K, Norouzi M, Bayat P, Karlikow M, Cicek S, Guo Y, Green AA, Pena L, Pardee K. JoVE 2022;(184). PMID: 35781278
Portable sample processing for molecular assays: application to Zika virus diagnostics
Narahari T, Dahmer J, Sklavounos A, Kim T, Satkauskas M, Clotea I, Ho M, Lamanna J, Dixon C, Rackus D, da Silva R, Pena L, Pardee K, Wheeler AR. Lab-on-a-Chip. 22: 1748-1763. (2022) PMID. 35357372
Multicenter international assessment of a SARS-CoV-2 RT-LAMP test for point of care clinical application
Lu S, Duplat D, Benitez-Bolivar P, León C, Villota SD, Veloz-Villavicencio E, Arévalo V, Jaenes K, Guo Y, Cicek S, Robinson L, Peidis P, Pearson JD, Woodgett J, Mazzulli T, Ponce P, Restrepo S, González JM, Bernal A, Guevara-Suarez M, Pardee K, Cevallos VE, González C, Bremner R. PLOS ONE. 17(5): e0268340. (2022) PMID. 35544541
Two Years into the COVID-19 Pandemic: Lessons Learned
da Silva SJR, do Nascimento JCF, Germano Mendes RP, Guarines KM, Targino Alves da Silva C, da Silva PG, de Magalhães JJF, Vigar JRJ, Silva-Júnior A, Kohl A, Pardee K, Pena L. ACS Infect Dis. 2022;8(9): 1758-1817. PMID. 35940589.
Logic Invades Cell-free Biosensing
Amalfitano E and Pardee K.
Nature Chemical Biology, Feb 2022.
A glucose meter interface for point-of-care gene circuit-based diagnostics
Amalfitano E, Karlikow M, Norouzi M, Jaenes K, Cicek S, Masum F, Sadat Mousavi P, Guo Y, Tang L, Sydor A, Ma D, Pearson JD, Trcka D, Pinette M, Ambagala A, Babiuk S, Pickering B, Wrana J, Bremner R, Mazzulli T, Sinton D, Brumell JH, Green AA, Pardee K. Nat Commun. 2021 Feb 1;12(1):724.
High-Efficiency Protection of Linear DNA in Cell-Free Extracts from Escherichia coli and Vibrio natriegens
Norouzi M, Panfilov S, Pardee K. ACS Synth Biol. 2021 Jul 16;10(7):1615-1624.
Decentralizing Cell-Free RNA Sensing With the Use of Low-Cost Cell Extracts
Arce A, Guzman Chavez F, Gandini C, Puig J, Matute T, Haseloff J, Dalchau N, Molloy J, Pardee K, Federici F. Front Bioeng Biotechnol. 2021 Aug 23;9:727584.
Development and validation of a one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) for rapid detection of ZIKV in patient samples from Brazil
da Silva SJR, Pardee K, Balasuriya UBR, Pena L. Sci Rep. 2021 Feb 18;11(1):4111.
Adaptive, diverse and de-centralized diagnostics are key to the future of outbreak response
Matthews Q, da Silva SJR, Norouzi M, Pena LJ, Pardee K.. BMC Biol. 2020 Oct 28;18(1):153.
Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19
da Silva SJR, Silva CTAD, Guarines KM, Mendes RPG, Pardee K, Kohl A, Pena L. ACS Infect Dis. 2020 Sep 11;6(9):2319-2336.
When robotics met fluidics
Zhong J, Riordon J, Wu TC, Edwards H, Wheeler AR, Pardee K, Aspuru-Guzik A, Sinton D.Lab Chip. 2020 Feb 21;20(4):709-716.
A Multiplexed, Electrochemical Interface for Gene Circuit-Based Sensors
Sadat Mousavi P, Smith SJ, Chen JB, Karlikow M, Tinafar A, Robinson C, Liu W, Ma D, Green AA, Kelley SO, Pardee K. Nat Chem. 2020 Jan;12(1):48-55.
Loop-Mediated Isothermal Amplification (LAMP) for the Diagnosis of Zika Virus: A Review
Silva SJRD, Pardee K, Pena L.. Viruses. 2019 Dec 23;12(1):19. doi: 10.3390/v12010019. PMID: 31877989; PMCID: PMC7019470.
Synthetic Biology Goes Cell-Free
Tinafar A, Jaenes K, Pardee K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019 Aug 8;17(1):64.
The Many Roads to an Ideal Paper-based Device
Karlikow M, Pardee K. Paper-Based Diagnostics. Online: Springer International Publishing;2018
Perspective: Solidifying the impact of cell-free synthetic biology through lyophilization
Pardee K. Biochem Eng J. 2018 Oct 15;138:91-97.
BioBits™ Explorer: A modular synthetic biology education kit
Huang A, Nguyen PQ, Stark JC, Takahashi MK, Donghia N, Ferrante T, Dy AJ, Hsu KJ, Dubner RS, Pardee K, Jewett MC, Collins JJ. Sci Adv. 2018 Aug 1;4(8):eaat5105.
BioBits™ Bright: A fluorescent synthetic biology education kit
Stark JC, Huang A, Nguyen PQ, Dubner RS, Hsu KJ, Ferrante TC, Anderson M, Kanapskyte A, Mucha Q, Packett JS, Patel P, Patel R, Qaq D, Zondor T, Burke J, Martinez T, Miller-Berry A, Puppala A, Reichert K, Schmid M, Brand L, Hill LR, Chellaswamy JF, Faheem N, Fetherling S, Gong E, Gonzalzles EM, Granito T, Koritsaris J, Nguyen B, Ottman S, Palffy C, Patel A, Skweres S, Slaton A, Woods T, Donghia N, Pardee K, Collins JJ, Jewett MC. Sci Adv. 2018 Aug 1;4(8):eaat5107
Portable, On-Demand Biomolecular Manufacturing
Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, Ferrante T, McSorley FR, Furuta Y, Vernet A, Lewandowski M, Boddy CN, Joshi NS, Collins JJ. Cell. 2016 Sep 22;167(1):248-259.e12.
Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components
Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. Cell. 2016 May 19;165(5):1255-1266.
Synthetic biology devices for in vitro and in vivo diagnostics
Slomovic S, Pardee K, Collins JJ. Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14429-35.
Paper-based synthetic gene networks
Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. Cell. 2014 Nov 6;159(4):940-54.
Deconstructing transcriptional heterogeneity in pluripotent stem cells
Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser A, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, Ferrante TC, Regev A, Daley GQ, Collins JJ. Nature. 2014 Dec 4;516(7529):56-61.
Gene networks of fully connected triads with complete auto-activation enable multistability and stepwise stochastic transitions
Faucon PC, Pardee K, Kumar RM, Li H, Loh YH, Wang X. PLoS One. 2014 Jul 24;9(7):e102873.
Nuclear Receptors: Small Molecule Sensors that Coordinate Growth, Metabolism and Reproduction
Pardee K, Necakov AS, Krause H. Subcell Biochem. 2011;52:123-53.
The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis
Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. Genes Dev. 2009 Dec 1;23(23):2711-6.
Nuclear receptors homo sapiens Rev-erbbeta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO
Marvin KA, Reinking JL, Lee AJ, Pardee K, Krause HM, Burstyn JN. Biochemistry. 2009 Jul 28;48(29):7056-71.
The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta
Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, Botchkarev A, Krause HM, Edwards A. PLoS Biol. 2009 Feb 24;7(2):e43.
The Drosophila nuclear receptor e75 contains heme and is gas responsive
Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, Krause HM. Cell. 2005 Jul 29;122(2):195-207.
Nuclear hormone receptors, metabolism, and aging: what goes around comes around. Transcription factors link lipid metabolism and aging-related processes
Pardee K, Reinking J, Krause H. Sci Aging Knowledge Environ. 2004 Nov 24;2004(47):re8
Nuclear Receptors: Small Molecule Sensors that Coordinate Growth, Metabolism and Reproduction
Pardee K, Necakov AS, Krause H. Subcell Biochem. 2011;52:123-53.
P1 Trisaccharide (Galalpha1,4Galbeta1,4GlcNAc) synthesis by enzyme glycosylation reactions using recombinant Escherichia coli
Liu Z, Lu Y, Zhang J, Pardee K, Wang PG. Appl Environ Microbiol. 2003 Apr;69(4):2110-5.
Structural proteomics: toward high-throughput structural biology as a tool in functional genomics
Yee A, Pardee K, Christendat D, Savchenko A, Edwards AM, Arrowsmith CH. Acc Chem Res. 2003 Mar;36(3):183-9.
Plant Virus Inhibitors from Marine Algae
Pardee K, Ellis P, Bouthillier M, Towers GHN, French CJ
Canadian Journal of Botany. 2004 Vol. 82 (3): 304-309.
Karlikow M, Amalfitano E, Yang X, Doucet J, Chapman A, Mousavi PS, Homme P, Sutyrina P, Chan W, Lemak S, Yakunin AF, Dolezal AG, Kelley S, Foster LJ, Harpur B, Pardee K. Nature Communications. 2023 Mar 17;14(1):1505. PMID: 36932065.
Clinical and Laboratory Diagnosis of Monkeypox (Mpox): Current Status and Future Directions
Ribeiro da Silva J, Kohl A, Pena L, Pardee K. iScience 2023. 2023 Jun 16;26(6):106759. PMID: 37206155.
Cell-Free Dot Blot: an Ultra-Low-Cost and Practical Immunoassay Platform for Detection of Anti-SARS-CoV-2 Antibodies in Human and Animal Sera
Norouzi M, Truong T, Jaenes K, Warner BM, Vendramelli R, Tierney K, Kobasa D, Tailor N, Plant P, Dos Santos C, Babiuk S, Ambagala A, Pardee K. Microbiol Spectr 2023 Jan 31:e0245722. PMID: 36719206.
Recent insights into SARS-CoV-2 omicron variant
Ribeiro da Silva J, Kohl A, Pena L, Pardee K. Reviews in Medical Virology. e2373. (2023) PMID: 35662313
Field Validation of the Performance of Paper-Based Tests for the Detection of the Zika And Chikungunya Viruses in Serum Samples
Karlikow M, Ribeiro J, Guo Y, Cicek S, Krokovski L, Homme P, Xiong Y, Xu T, Calderon M, Camacho S, Ma D, Nayane B, Sutyrina P, Ferrante T, Benitez D, Tamayo V, Jaenes K, Rackus D, Collins J, Castellanos J, Cevallos V, Green AA, Ayres C, Pena L, Pardee K. Nature Biomedical Engineering, Mar 2022.
Toward Mail-in-Sensors for SARS-CoV-2 Detection: Interfacing Gel Switch Resonators with Cell-Free Toehold Switches
Carr AR, Dopp JL, Wu K, Sadat Mousavi P, Jo YR, McNeley CE, Lynch ZT, Pardee K, Green AA, Reuel NF. T. ACS Sensors. 7, 3, 806–815 (2022) PMID. 35254055.
From Design to Implementation: The Development and Patient Validation of Paper-Based Toehold Switch-Based Diagnostics
Jaenes K, Ribeiro J, Vigar J, Wu K, Norouzi M, Bayat P, Karlikow M, Cicek S, Guo Y, Green AA, Pena L, Pardee K. JoVE 2022;(184). PMID: 35781278
Portable sample processing for molecular assays: application to Zika virus diagnostics
Narahari T, Dahmer J, Sklavounos A, Kim T, Satkauskas M, Clotea I, Ho M, Lamanna J, Dixon C, Rackus D, da Silva R, Pena L, Pardee K, Wheeler AR. Lab-on-a-Chip. 22: 1748-1763. (2022) PMID. 35357372
Multicenter international assessment of a SARS-CoV-2 RT-LAMP test for point of care clinical application
Lu S, Duplat D, Benitez-Bolivar P, León C, Villota SD, Veloz-Villavicencio E, Arévalo V, Jaenes K, Guo Y, Cicek S, Robinson L, Peidis P, Pearson JD, Woodgett J, Mazzulli T, Ponce P, Restrepo S, González JM, Bernal A, Guevara-Suarez M, Pardee K, Cevallos VE, González C, Bremner R. PLOS ONE. 17(5): e0268340. (2022) PMID. 35544541
Two Years into the COVID-19 Pandemic: Lessons Learned
da Silva SJR, do Nascimento JCF, Germano Mendes RP, Guarines KM, Targino Alves da Silva C, da Silva PG, de Magalhães JJF, Vigar JRJ, Silva-Júnior A, Kohl A, Pardee K, Pena L. ACS Infect Dis. 2022;8(9): 1758-1817. PMID. 35940589.
Logic Invades Cell-free Biosensing
Amalfitano E and Pardee K.
Nature Chemical Biology, Feb 2022.
A glucose meter interface for point-of-care gene circuit-based diagnostics
Amalfitano E, Karlikow M, Norouzi M, Jaenes K, Cicek S, Masum F, Sadat Mousavi P, Guo Y, Tang L, Sydor A, Ma D, Pearson JD, Trcka D, Pinette M, Ambagala A, Babiuk S, Pickering B, Wrana J, Bremner R, Mazzulli T, Sinton D, Brumell JH, Green AA, Pardee K. Nat Commun. 2021 Feb 1;12(1):724.
High-Efficiency Protection of Linear DNA in Cell-Free Extracts from Escherichia coli and Vibrio natriegens
Norouzi M, Panfilov S, Pardee K. ACS Synth Biol. 2021 Jul 16;10(7):1615-1624.
Decentralizing Cell-Free RNA Sensing With the Use of Low-Cost Cell Extracts
Arce A, Guzman Chavez F, Gandini C, Puig J, Matute T, Haseloff J, Dalchau N, Molloy J, Pardee K, Federici F. Front Bioeng Biotechnol. 2021 Aug 23;9:727584.
Development and validation of a one-step reverse transcription loop-mediated isothermal amplification (RT-LAMP) for rapid detection of ZIKV in patient samples from Brazil
da Silva SJR, Pardee K, Balasuriya UBR, Pena L. Sci Rep. 2021 Feb 18;11(1):4111.
Adaptive, diverse and de-centralized diagnostics are key to the future of outbreak response
Matthews Q, da Silva SJR, Norouzi M, Pena LJ, Pardee K.. BMC Biol. 2020 Oct 28;18(1):153.
Clinical and Laboratory Diagnosis of SARS-CoV-2, the Virus Causing COVID-19
da Silva SJR, Silva CTAD, Guarines KM, Mendes RPG, Pardee K, Kohl A, Pena L. ACS Infect Dis. 2020 Sep 11;6(9):2319-2336.
When robotics met fluidics
Zhong J, Riordon J, Wu TC, Edwards H, Wheeler AR, Pardee K, Aspuru-Guzik A, Sinton D.Lab Chip. 2020 Feb 21;20(4):709-716.
A Multiplexed, Electrochemical Interface for Gene Circuit-Based Sensors
Sadat Mousavi P, Smith SJ, Chen JB, Karlikow M, Tinafar A, Robinson C, Liu W, Ma D, Green AA, Kelley SO, Pardee K. Nat Chem. 2020 Jan;12(1):48-55.
Loop-Mediated Isothermal Amplification (LAMP) for the Diagnosis of Zika Virus: A Review
Silva SJRD, Pardee K, Pena L.. Viruses. 2019 Dec 23;12(1):19. doi: 10.3390/v12010019. PMID: 31877989; PMCID: PMC7019470.
Synthetic Biology Goes Cell-Free
Tinafar A, Jaenes K, Pardee K. Synthetic Biology Goes Cell-Free. BMC Biol. 2019 Aug 8;17(1):64.
The Many Roads to an Ideal Paper-based Device
Karlikow M, Pardee K. Paper-Based Diagnostics. Online: Springer International Publishing;2018
Perspective: Solidifying the impact of cell-free synthetic biology through lyophilization
Pardee K. Biochem Eng J. 2018 Oct 15;138:91-97.
BioBits™ Explorer: A modular synthetic biology education kit
Huang A, Nguyen PQ, Stark JC, Takahashi MK, Donghia N, Ferrante T, Dy AJ, Hsu KJ, Dubner RS, Pardee K, Jewett MC, Collins JJ. Sci Adv. 2018 Aug 1;4(8):eaat5105.
BioBits™ Bright: A fluorescent synthetic biology education kit
Stark JC, Huang A, Nguyen PQ, Dubner RS, Hsu KJ, Ferrante TC, Anderson M, Kanapskyte A, Mucha Q, Packett JS, Patel P, Patel R, Qaq D, Zondor T, Burke J, Martinez T, Miller-Berry A, Puppala A, Reichert K, Schmid M, Brand L, Hill LR, Chellaswamy JF, Faheem N, Fetherling S, Gong E, Gonzalzles EM, Granito T, Koritsaris J, Nguyen B, Ottman S, Palffy C, Patel A, Skweres S, Slaton A, Woods T, Donghia N, Pardee K, Collins JJ, Jewett MC. Sci Adv. 2018 Aug 1;4(8):eaat5107
Portable, On-Demand Biomolecular Manufacturing
Pardee K, Slomovic S, Nguyen PQ, Lee JW, Donghia N, Burrill D, Ferrante T, McSorley FR, Furuta Y, Vernet A, Lewandowski M, Boddy CN, Joshi NS, Collins JJ. Cell. 2016 Sep 22;167(1):248-259.e12.
Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components
Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Ferrante T, Ma D, Donghia N, Fan M, Daringer NM, Bosch I, Dudley DM, O'Connor DH, Gehrke L, Collins JJ. Cell. 2016 May 19;165(5):1255-1266.
Synthetic biology devices for in vitro and in vivo diagnostics
Slomovic S, Pardee K, Collins JJ. Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14429-35.
Paper-based synthetic gene networks
Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. Cell. 2014 Nov 6;159(4):940-54.
Deconstructing transcriptional heterogeneity in pluripotent stem cells
Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser A, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, Ferrante TC, Regev A, Daley GQ, Collins JJ. Nature. 2014 Dec 4;516(7529):56-61.
Gene networks of fully connected triads with complete auto-activation enable multistability and stepwise stochastic transitions
Faucon PC, Pardee K, Kumar RM, Li H, Loh YH, Wang X. PLoS One. 2014 Jul 24;9(7):e102873.
Nuclear Receptors: Small Molecule Sensors that Coordinate Growth, Metabolism and Reproduction
Pardee K, Necakov AS, Krause H. Subcell Biochem. 2011;52:123-53.
The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis
Horner MA, Pardee K, Liu S, King-Jones K, Lajoie G, Edwards A, Krause HM, Thummel CS. Genes Dev. 2009 Dec 1;23(23):2711-6.
Nuclear receptors homo sapiens Rev-erbbeta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO
Marvin KA, Reinking JL, Lee AJ, Pardee K, Krause HM, Burstyn JN. Biochemistry. 2009 Jul 28;48(29):7056-71.
The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta
Pardee KI, Xu X, Reinking J, Schuetz A, Dong A, Liu S, Zhang R, Tiefenbach J, Lajoie G, Plotnikov AN, Botchkarev A, Krause HM, Edwards A. PLoS Biol. 2009 Feb 24;7(2):e43.
The Drosophila nuclear receptor e75 contains heme and is gas responsive
Reinking J, Lam MM, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, Krause HM. Cell. 2005 Jul 29;122(2):195-207.
Nuclear hormone receptors, metabolism, and aging: what goes around comes around. Transcription factors link lipid metabolism and aging-related processes
Pardee K, Reinking J, Krause H. Sci Aging Knowledge Environ. 2004 Nov 24;2004(47):re8
Nuclear Receptors: Small Molecule Sensors that Coordinate Growth, Metabolism and Reproduction
Pardee K, Necakov AS, Krause H. Subcell Biochem. 2011;52:123-53.
P1 Trisaccharide (Galalpha1,4Galbeta1,4GlcNAc) synthesis by enzyme glycosylation reactions using recombinant Escherichia coli
Liu Z, Lu Y, Zhang J, Pardee K, Wang PG. Appl Environ Microbiol. 2003 Apr;69(4):2110-5.
Structural proteomics: toward high-throughput structural biology as a tool in functional genomics
Yee A, Pardee K, Christendat D, Savchenko A, Edwards AM, Arrowsmith CH. Acc Chem Res. 2003 Mar;36(3):183-9.
Plant Virus Inhibitors from Marine Algae
Pardee K, Ellis P, Bouthillier M, Towers GHN, French CJ
Canadian Journal of Botany. 2004 Vol. 82 (3): 304-309.